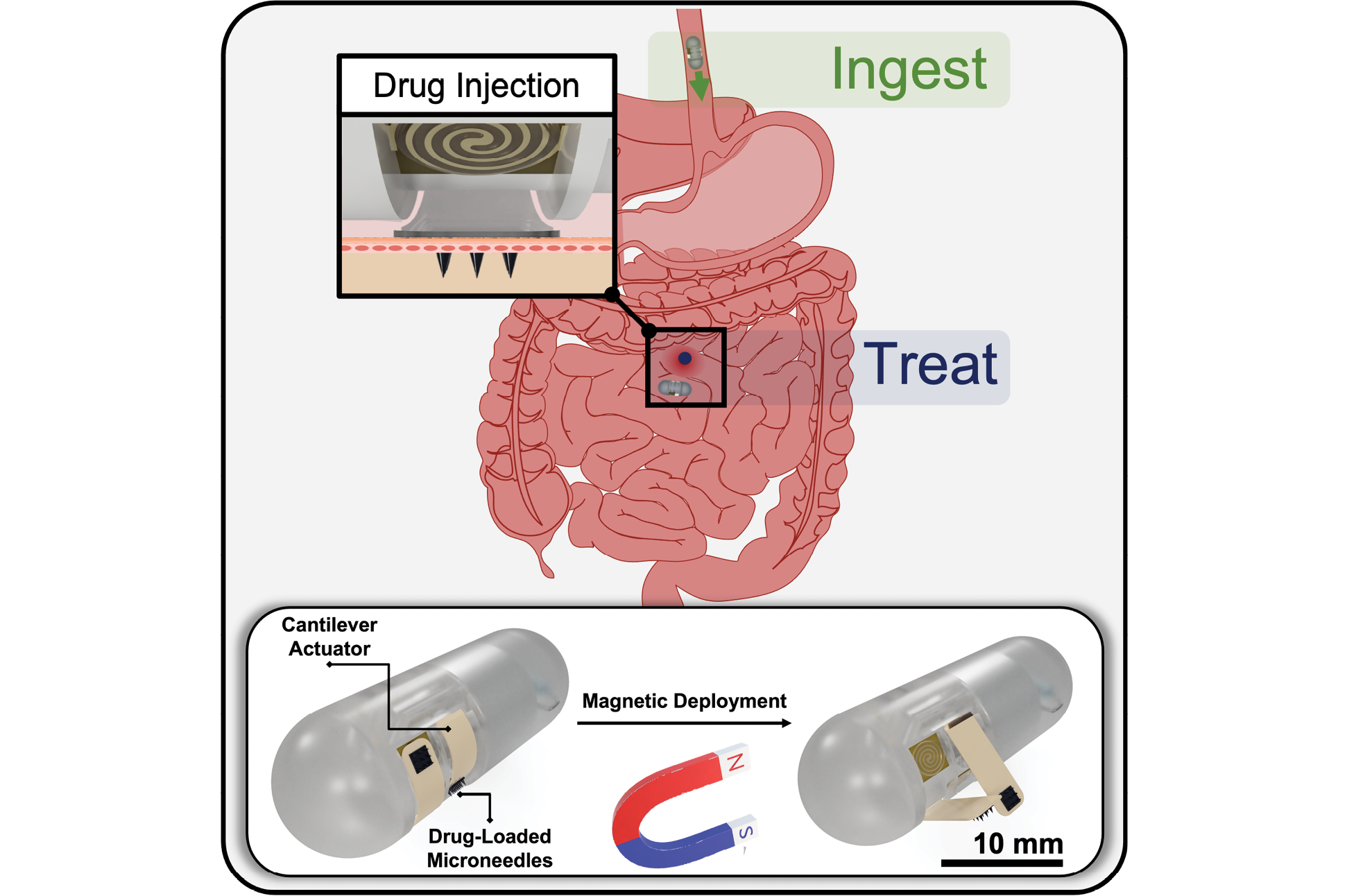
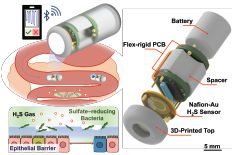
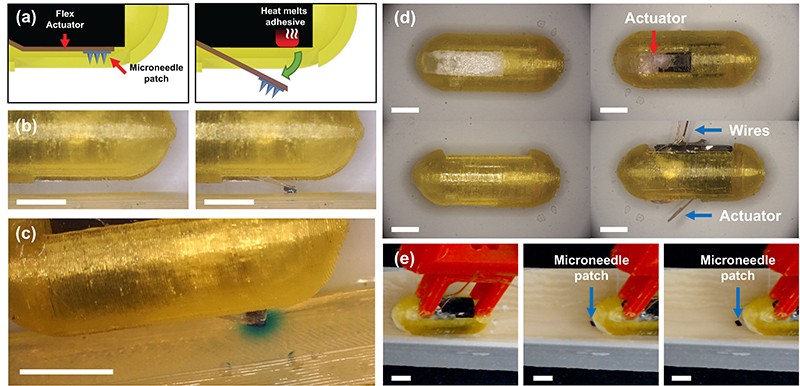
News Story
Symptoms all in your head—or in your gut? Maybe a little of both.
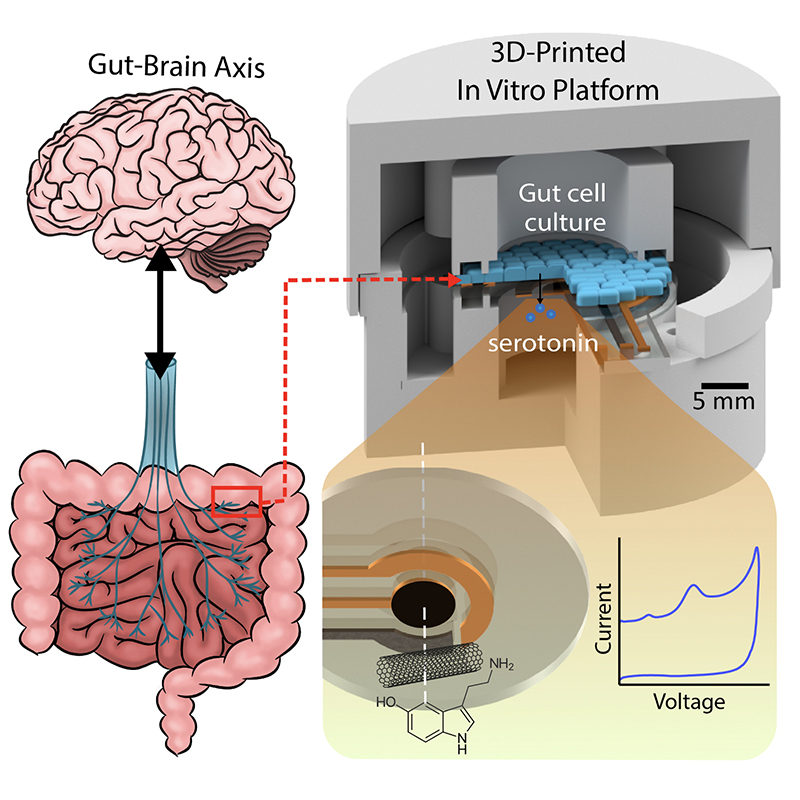
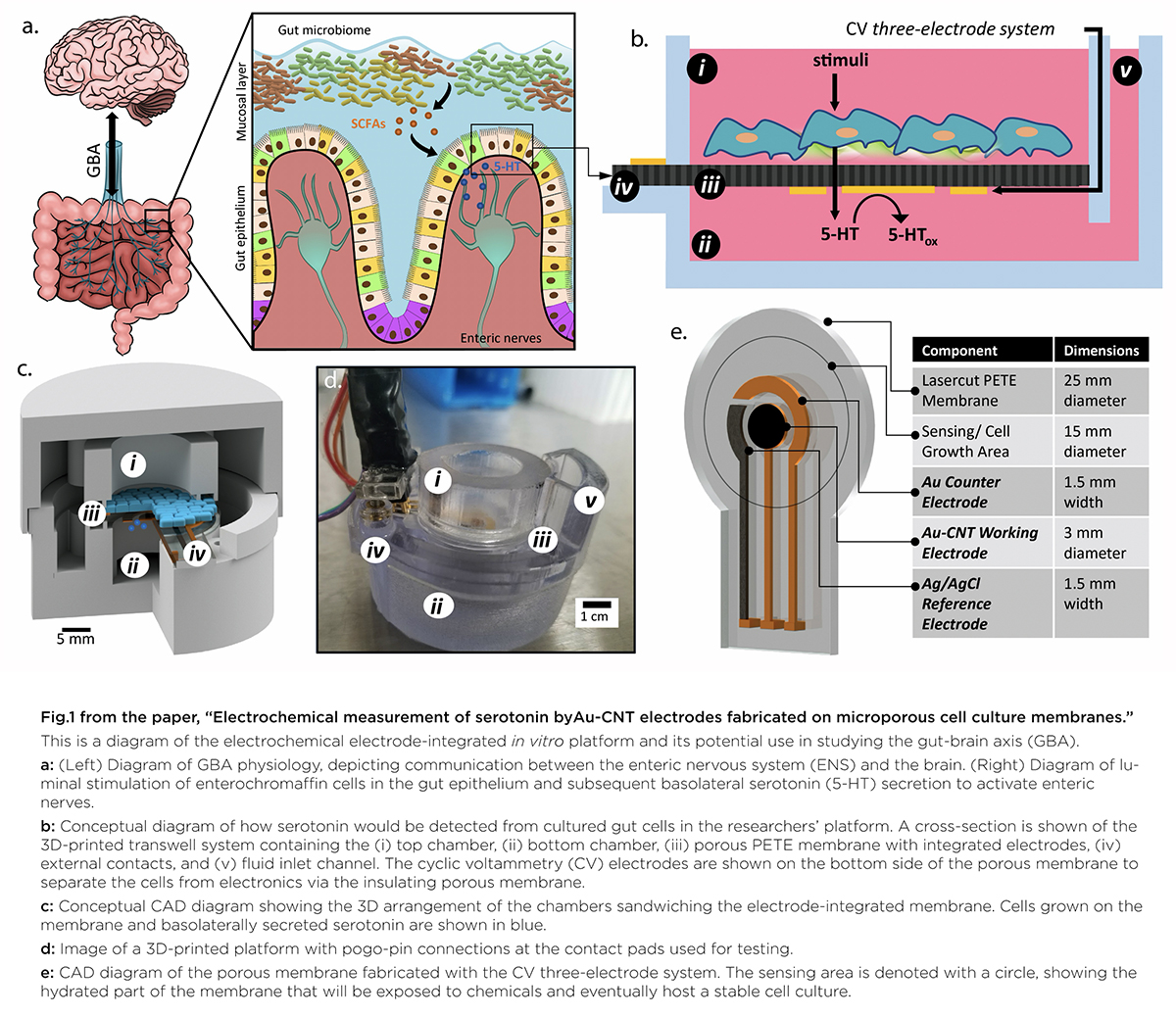
A graphical abstract of the gut-brain axis (left) and the 3D-printed in vitro platform (right).
Anyone who has ever experienced “butterflies in the stomach” before giving a big presentation won’t be surprised to learn there is an actual physical connection between their gut and their brain. Neuroscientists and medical professionals call this the “gut-brain-axis” (GBA). A better understanding of the GBA could lead to treatments and cures for neurological mood disorders like depression and anxiety, as well as for a range of chronic auto-immune inflammatory diseases like irritable bowel syndrome (IBS) and rheumatoid arthritis (RA).
The problem is that up until now “butterflies” have been all medical doctors have had to work with in detecting these GBA-related disorders. Even today these diseases are primarily diagnosed by patients’ own reports of their symptoms. Finding “biomarkers”—objective measurements of a substance in the body that indicates a disorder’s presence—could dramatically improve diagnosis and take an enormous burden off patients to correctly identify their own symptoms.
Scientists suspect the chemical neurotransmitter serotonin is the biomarker for a range of GBA disorders. Serotonin spurs the nervous system into action via the vagus nerve, the physical connector between the brain and the colon. Generated deep within the lining of the gut, serotonin ultimately influences everything from mood and emotions to sleep, digestion and the secretion of hormones. Its production is in some way affected by the bacterial “microbiome” present in this environment. Researchers hope that creating tools to analyze serotonin’s production and dysfunction in the gut microbiome will help unlock the mysteries of GBA-related disorders.
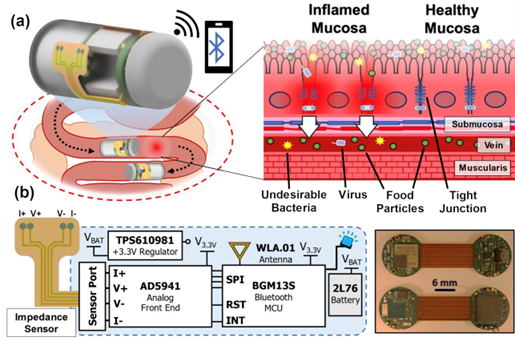
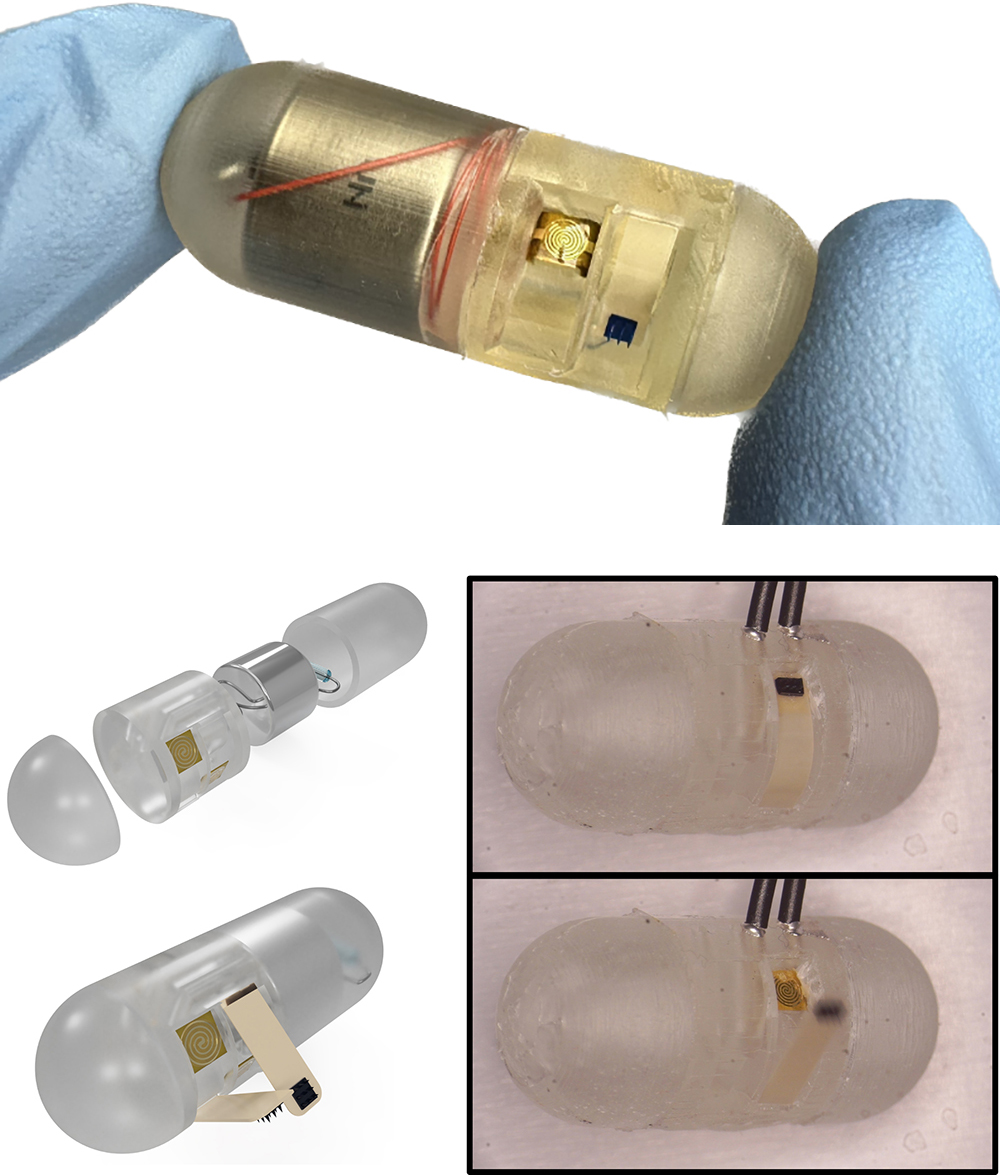
With $1million in National Science Foundation funding, University of Maryland (UMD) engineers, neuroscientists, microbiologists and physicists have made significant progress in developing a platform that can monitor and model the real-time processing of gut microbiome serotonin activity. Their goal is to one day package the platform into an ingestible capsule capable of detecting, treating and monitoring GBA diseases.
Converging disciplines is key, says Professor Reza Ghodssi, the project’s principal investigator. “We are converging neuroscience, molecular signaling, and micro-nano devices and systems. This enables us to measure and investigate data at the interface of each junction of a simulated GBA platform—cell to cell, cell to molecule, molecule to nerve—and develop engineering methodologies to analyze and interpret it.”
The work builds on ingestible medical device expertise that has been developed in the UMD MEMS Sensors and Actuators Laboratory, the Fischell Department of Bioengineering, and the Brain and Behavior Initiative.
Three new published papers detail the progress in detecting serotonin, assessing its neurological effects, and sensing minute changes to the gut epithelium.
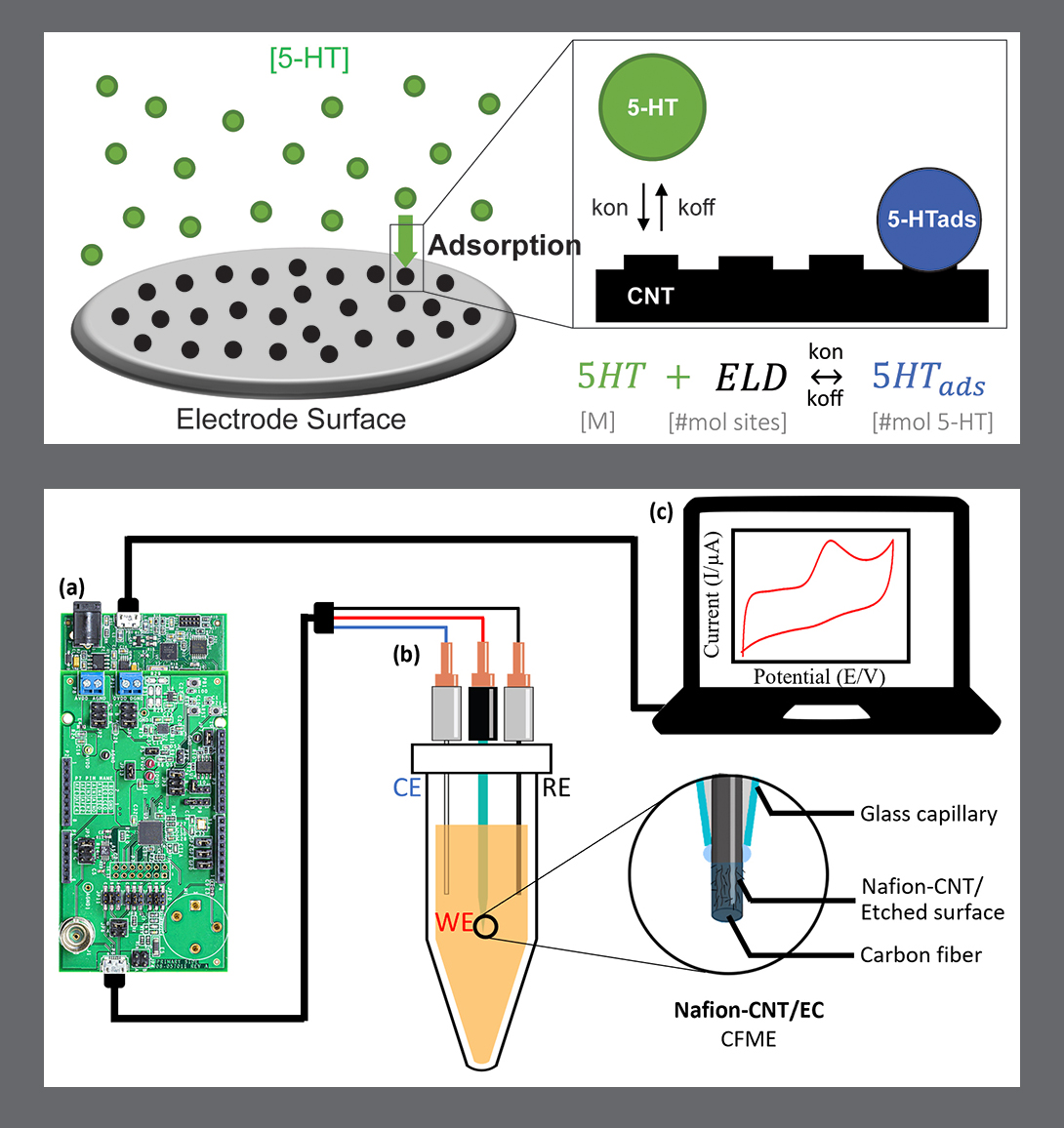
In “Electrochemical Measurement of Serotonin by Au-CNT Electrodes Fabricated on Porous Cell Culture Membranes,” the team developed a platform that provides access to the specific site of serotonin production, important because serotonin is secreted from the bottoms of cells. An innovative porous membrane with an integrated serotonin sensor on which a model of the gut lining can be grown allowed researchers to access both top and bottom sides of the cell culture.
The paper was published online Sept. 7, 2020 in the Nature journal, Microsystems and Nanoengineering. The authors are Bioengineering Ph.D. student Ashley Chapin, former ISR postdoctoral researcher Pradeep Ramiah Rajasekaran, alumnus David N. Quan (BioE Ph.D. 2015), Professor Liangbing Hu (MSE/MEII), Associate Professor Jens Herberholz (Psychology/NACS), Professor William Bentley (BioE/Fischell Institute/IBBR), and Professor Reza Ghodssi (ECE/ISR).
Using metal deposition, they patterned a three-electrode system directly on a porous cell culture membrane suspended within a custom 3D-printed housing. Cells can be grown on the top of the membrane with the serotonin sensor oriented on the bottom for direct detection. The team then enhanced the sensitivity of serotonin detection by increasing the electrode effective surface area, drop-casting a small volume of carbon nanotubes on the electrode surface. Prepared solutions of serotonin were detectable well within the expected physiological concentration range.
The work is the first to demonstrate a feasible method for detecting redox molecules—such as serotonin—directly on a porous and flexible cell culture substrate. It grants superior access to cell-released molecules and creates a controllable model gut environment without resorting to invasive procedures on humans or animals.
The team’s second paper, “A Hybrid Biomonitoring System for Gut-Neuron Communication,” builds on the findings of the first: the researchers developed the serotonin measuring platform further so it could assess serotonin’s neurological effects. By adding and integrating a dissected crayfish nerve model with the gut lining model, the team created a gut-neuron interface that can electrophysiologically assess nerve response to the electrochemically detected serotonin. This advance enables the study of molecular signaling between gut and nerve cells, making possible real-time monitoring of both GBA tissues for the first time.
The paper was published online in the June 2020 IEEE Journal of Microelectromechanical Systems. It was written by Chapin, Electrical and Computer Engineering Ph.D. student Jinjing Han, Neuroscience and Cognitive Science Ph.D. student Ta-Wen Ho, Herberholz and Ghodssi.
| View a larger version of this diagram |
Finally, the concept, design and use for the entire biomonitoring platform is described in a third paper, “3D Printed Electrochemical Sensor Integrated Transwell Systems,” published online Oct. 5, 2020 in the Nature journal Microsystems and Nanoengineering. The paper was written by Rajasekaran, Chapin, Quan, Herberholz, Bentley and Ghodssi.
This paper delves into the development of the 3D-printed housing, the maintenance of a healthy lab-on-a-chip gut cell culture, and the evaluation of the two types of sensors integrated on the cell culture membrane. The dual sensors are particularly important because they provide feedback about multiple components of the system—namely, the portions that model the gut lining’s permeability (a strong indicator of disease) and its serotonin release (a measure of communication with the nervous system). Alongside the electrochemical sensor—evaluated using a standard redox molecule ferrocene dimethanol—an impedance sensor was used to monitor cell growth and coverage over the membrane. Using both these sensors would allow monitoring of a gut cell culture under various environmental and dietary conditions. It also would enable researchers to evaluate changes to barrier permeability (a strong indicator of disease), and serotonin release (a measure of communication with the nervous system).
“These works represent a big step forward in our understanding of the gut/brain axis,” says Cornell University’s John March, Chair of the Department of Biological and Environmental Engineering. “One of the limitations of this field is the inability to perform highly controlled experiments in a ‘close to in vivo’ system. These papers provide ways around this problem with simple, elegant experiments that are highly accessible. I expect these will be used frequently.”
Because the engineering aspects of the platform are well underway, the researchers are working towards culturing multi-tissue interfaces with the help of Jay Pasricha and Subhash Kulkarni at Johns Hopkins University. Eventually multiple platforms will be created, each colonized with a different combination of gut bacteria, to measure the neurophysiological effects of serotonin production in varying microbiome environments.
With this information, Professor Wolfgang Losert (Physics/IPST/IREAP) will lead a machine learning effort to process the sensor data through a computer model that can simulate the outcomes of the different microbiomes. This will provide the clearest picture yet of how a system as complex and individually unique as the gut microbiome affects both gut and brain health. It also may help researchers better understand the connection between nutrition and mental health.
“Understanding biology at the level of whole organisms is a frontier in biology, and essential to forming a basis for precision medicine,” says the University of California, Berkeley’s Amy Herr, the John D. & Catherine T. MacArthur Professor of Bioengineering. “By harnessing hallmarks of engineering—integrated, systems-level design—the new research from the Ghodssi-Bentley-Herberholz team presents an integrated approach to elegantly perturb and then probe the electrons and molecules that are key conduits of information flow in whole organisms.”
Published October 14, 2020